Agdia, Inc. (Elkhart, IN) is happy to announce the commercialization of a rapid, user-friendly, DNA-based assay, on their AmplifyRP® XRT platform, for the detection of Beet curly top virus.
Curly top disease affects numerous commercially important hosts, including common bean (Phaseolus vulgaris), cucumber (Cucumis sativus), industrial hemp (Cannabis sativa), pepper (Capsicum annuum), potato (Solanum tuberosum), spinach (Spinacia oleracea), squash and pumpkin (Cucurbita pepo), sugar and table beet (Beta vulgaris) and tomato (Solanum lycopersicum). This disease is caused by Beet curly top virus (BCTV), a Curtovirus (family Geminiviridae), existing as a complex of strains differentiated genotypically, causing symptomology on the hosts mentioned above. Additionally, strains of BCTV are known to infect more than 300 species of plants in no fewer than 44 families, many of which are asymptomatic, weedy hosts.
Symptomology of Beet curly top virus was first observed in the late 19th century in the western U.S. on sugar beets. It was, however, not recognized as being caused by a specific pathogen until 1915, when leafhopper transmission was proven, and viral etiology was proposed. Since then, BCTV has spread throughout North America where hosts are cultivated, including several states in the American West and Southwest, southwestern Canada and Mexico. Furthermore, BCTV has been identified in parts of South America and several countries in the Mediterranean basin. All strains of BCTV are considered pathogens of quarantine importance in Canada, Israel, Mexico and the European Union.
Beet curly top virus is transmitted efficiently by the beet leafhopper, Circulifer tenellus (Order Hemiptera), in a persistent circulative manner. The virus can be acquired within minutes of feeding, and insects are known to remain viruliferous for up to a month. Beet curly top virus is phloem-limited, and the leafhopper must feed on infected phloem to acquire and transmit the virus to healthy plants. Circulifer tenellus is the only known vector in North America; however, in Europe, C. opacipennis is also known to vector the virus. The robust dynamics of the host-virus-vector relationship facilitate epidemics in parts of the world where leafhopper populations are high. Furthermore, the movement of infected propagative materials can spread the virus across great distances. Mechanical transmission through infected plant sap has been accomplished under experimental conditions. Nevertheless, this scenario is not thought to contribute to the epidemiology of naturally occurring curly top infections. Seed transmission of BCTV is not known to occur in the host species listed above.

Symptoms of curly top disease vary according to host and are typically more severe when plants are infected at earlier growth stages; many plants die before reaching maturity. Symptoms of curly top include stunted and distorted plant growth; leaf curling, crumpling, yellowing, vein swelling and distortion; and necrosis and hyperplasia of the phloem (figure 1). On beets, phloem tissue becomes necrotic, and exudate appears on the leaf surface. On tomato and pepper, fruit set is greatly diminished, and fruit that does form ripens prematurely. Furthermore, veins become purple.
Industrial hemp has reemerged as an important crop within several U.S. states, due to federal legalization and the demand for fiber, seed and cannabidiol. As production has increased, the list of disease organisms infecting this crop has grown to include several fungal, bacterial, viral and viroid pathogens. Beet curly top virus has been confirmed infecting industrial hemp and appears to be widespread on this crop throughout regions where vectors are present. There is a scarcity of research on this specific pathosystem; however, the understanding of the epidemiology of BCTV on cannabis is burgeoning, along with the crop. Symptoms of infection on industrial hemp include stunting leaf deformation and chlorosis (figure 2).

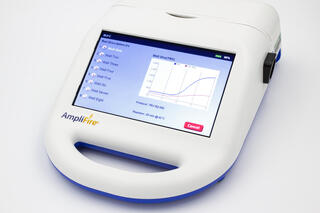
Agdia’s new AmplifyRP® XRT assay for detection BCTV is based on recombinase polymerase amplification (RPA). This technology promotes the rapid amplification and detection of nucleic acid targets, DNA or RNA, while maintaining a single operating temperature of 39 – 42 °C. The AmplifyRP® XRT products achieve target sensitivity and specificity comparable to PCR, while having clear advantages over the lab-based technology. AmplifyRP® XRT products do not require a nucleic acid purification step; crude sample extracts are prepared using a simple extraction buffer and tested directly. This makes the testing process simple and saves the end user valuable time. Furthermore, this facilitates the implementation of this technology at remote locations with limited resources. When paired with Agdia’s AmpliFire® isothermal fluorometer (figure 3), the XRT system is a rapid, user-friendly tool that can be implemented in the field or the lab by personnel with limited experience in molecular diagnostics.
Agdia states their assay was screened against a diverse collection of confirmed strains, including those infecting beets, industrial hemp, peppers and tomatoes, detecting all true positives. Furthermore, no cross-reactivity was observed with an extensive panel of viral and viroid pathogens, including Alfalfa mosaic virus, Cucumber mosaic virus, Hop latent viroid, Hop stunt viroid, Tobacco mosaic virus, Tobacco ringspot virus, Tomato brown rugose fruit virus, Tomato mosaic virus, Tomato ringspot virus, Tomato spotted wilt virus and Tomato yellow leaf curl virus. Sensitivity for this assay is greater than that observed with the published RT-qPCR assay and conventional RT-PCR assay to which it was compared. This product was developed to test leaf, stem and petiole tissue.
The introduction of this product brings Agdia’s catalog to 25 assays on the AmplifyRP® platform. High levels of market demand for field-deployable, plant pathogen detection products have driven this output, and Agdia maintains they will continue to expand their product offerings.



